Databases… a whole ‘big thing’ in the bioinformatics field. Databases are the “backbone” of all the public data banks of course, but can be very useful for many other aspects. Even our scaffolding program uses a database for fast data retrieval.
Databases stores data, as a very simple “text file”. But they have a powerful engine to quickly retrieve data using selected criteria. Consider our “pick primer” program: it takes a while to take the sequence of both contigs because it has to parse the file containing all of them. If we decided to store all sequences into a database we could perform a simple query to get them.
This blog is powered by a quite complex database.
What a database is
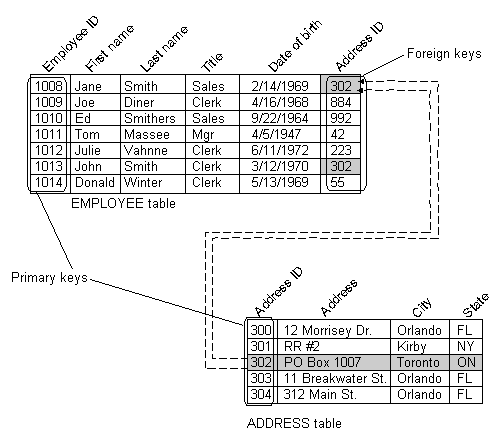
In very simple term a “database” is a collection of data, but it has a very precise structure: a database is a collection of tables, and each table has a set of columns called fields. Each row represent a single “item” and it’s called record. The picture below shows an example of a table (skip last field for now).

This figure shows a table representation of an EMPLOYEE table in a database, identifying the records and fields in the table. Each record in the table contains the following fields: Employee ID, First name, Last name, Title, Date of birth, Address ID.
Your mobile phone’s address book is a simple database consisting of a single table. Fields are Name, Surname, Phone, Email… and each contact is a record of the database.
Consider these two statements:
- Get me contacts whose surname starts for “Smi” (and sort them by surname first, then by first name)
- Get me contacts whose birthday is today
- Tell me how many contacts live in Padua
- Tell me the average length of the surname of my contacts
These are simple example queries.
Relational databases: tables falling in love
Sometimes with a single table we can do a lot, but relational databases interconnecting their tables are much better!
Consider you want to keep data about the SOLiD sequencer. Each sample has:
- a name (eg: “Grape leaf”)
- it belongs to a run (a run has many samples loaded into it)
- a library type (eg: “RNA Paired end”)
- a technician processing it (eg: “Micky”)
- a bioinfomatician analyzing it (eg: “Chuck Norris”)
- and many other fields of course!
- Name: Grape leaf
- Run: 193
- Processed by (name): Micky
- Processed by (mail): micky@mouse.com
MySQL
- Create databases and tables
- Insert records to a table
- Update a record in a table
- Query the table!
Source: http://docs.oracle.com/cd/A97329_03/toplink.903/b10061/concepts.htm