During our first lesson we had a brief overview of the sequencing strategy used for Nannochloropsis.
First, a whole genome shotgun approach was used to have a first draft (see picture below). In particular we used the Roche 454 machine that provides reads ~500bp long, and we assembled them with the Newbler package.
As we saw both read length and sequence coverage affects the quality of the assembly. In particular repeated regions make the assembly program to “break” the sequence. This happens if the length of the repeated region is longer than the single fragments (reads) sequenced. Repeated regions collapse in the same contig, that will have a higher coverage (approximately n-times the average, where n is the number of repeats in the genome).
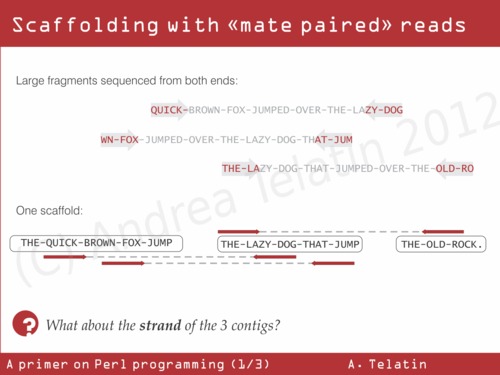
As we can’t have longer reads, we can use some molecular biology to put contigs in the correct order. If we fragment genomic DNA in large fragments (some kilobases, but anyway longer than common repeats) and we sequence both ends of each molecule, we can then align the two paired sequences against assembled contigs. Knowing the average distance between the two sequences… we can perform genome scaffolding (as depicted below).